Creating Publication-Quality Graphics with ggplot2
Last updated on 2026-03-03 | Edit this page
Estimated time: 80 minutes
Overview
Questions
- How can I create publication-quality graphics in R?
Objectives
- To be able to use ggplot2 to generate publication-quality graphics.
- To apply geometry, aesthetic, and statistics layers to a ggplot plot.
- To manipulate the aesthetics of a plot using different colors, shapes, and lines.
- To improve data visualization through transforming scales and paneling by group.
- To save a plot created with ggplot to disk.
Plotting our data is one of the best ways to quickly explore it and the various relationships between variables.
There are three main plotting systems in R, the base plotting system, the lattice package, and the ggplot2 package.
Today we’ll be learning about the ggplot2 package, because it is the most effective for creating publication-quality graphics.
ggplot2 is built on the grammar of graphics, the idea that any plot can be built from the same set of components: a data set, mapping aesthetics, and graphical layers:
Data sets are the data that you, the user, provide.
Mapping aesthetics are what connect the data to the graphics. They tell ggplot2 how to use your data to affect how the graph looks, such as changing what is plotted on the X or Y axis, or the size or color of different data points.
Layers are the actual graphical output from ggplot2. Layers determine what kinds of plot are shown (scatterplot, histogram, etc.), the coordinate system used (rectangular, polar, others), and other important aspects of the plot. The idea of layers of graphics may be familiar to you if you have used image editing programs like Photoshop, Illustrator, or Inkscape.
Let’s start off building an example using the gapminder data from
earlier. The most basic function is ggplot, which lets R
know that we’re creating a new plot. Any of the arguments we give the
ggplot function are the global options for the
plot: they apply to all layers on the plot.
R
library("ggplot2")
ggplot(data = gapminder)

Here we called ggplot and told it what data we want to
show on our figure. This is not enough information for
ggplot to actually draw anything. It only creates a blank
slate for other elements to be added to.
Now we’re going to add in the mapping aesthetics
using the aes function. aes tells
ggplot how variables in the data map to
aesthetic properties of the figure, such as which columns of
the data should be used for the x and
y locations.
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp))
Here we told ggplot we want to plot the “gdpPercap”
column of the gapminder data frame on the x-axis, and the “lifeExp”
column on the y-axis. Notice that we didn’t need to explicitly pass
aes these columns
(e.g. x = gapminder[, "gdpPercap"]), this is because
ggplot is smart enough to know to look in the
data for that column!
The final part of making our plot is to tell ggplot how
we want to visually represent the data. We do this by adding a new
layer to the plot using one of the
geom functions.
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point()
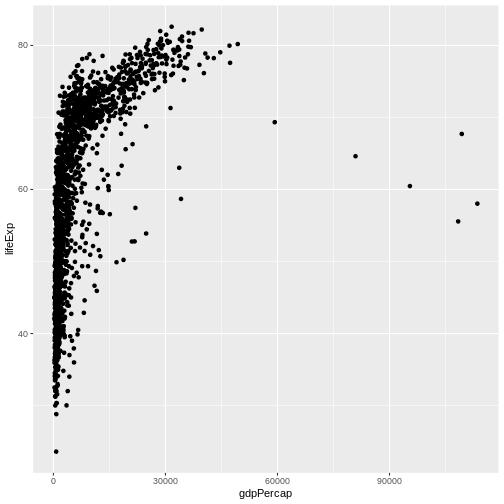
Here we used geom_point, which tells ggplot
we want to visually represent the relationship between
x and y as a scatterplot of
points.
Challenge 1
Modify the example so that the figure shows how life expectancy has changed over time:
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) + geom_point()
Hint: the gapminder dataset has a column called “year”, which should appear on the x-axis.
Here is one possible solution:
R
ggplot(data = gapminder, mapping = aes(x = year, y = lifeExp)) + geom_point()
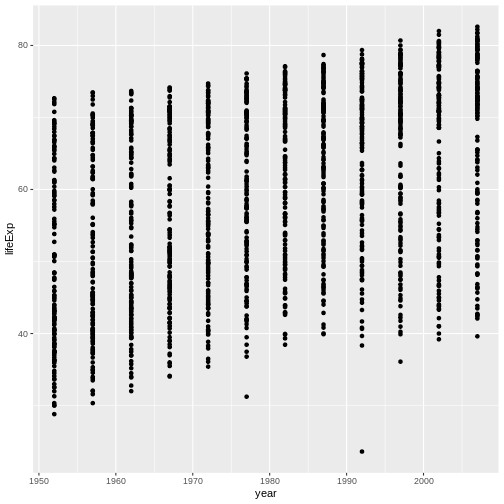
Challenge 2
In the previous examples and challenge we’ve used the
aes function to tell the scatterplot geom
about the x and y locations of each
point. Another aesthetic property we can modify is the point
color. Modify the code from the previous challenge to
color the points by the “continent” column. What trends
do you see in the data? Are they what you expected?
The solution presented below adds color=continent to the
call of the aes function. The general trend seems to
indicate an increased life expectancy over the years. On continents with
stronger economies we find a longer life expectancy.
R
ggplot(data = gapminder, mapping = aes(x = year, y = lifeExp, color=continent)) +
geom_point()
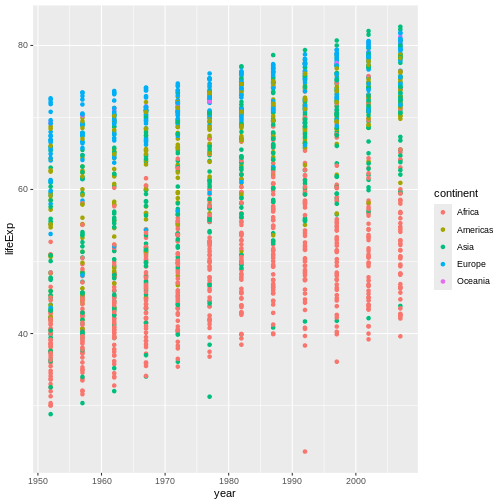
Layers
Using a scatterplot probably isn’t the best for visualizing change
over time. Instead, let’s tell ggplot to visualize the data
as a line plot:
R
ggplot(data = gapminder, mapping = aes(x=year, y=lifeExp, color=continent)) +
geom_line()
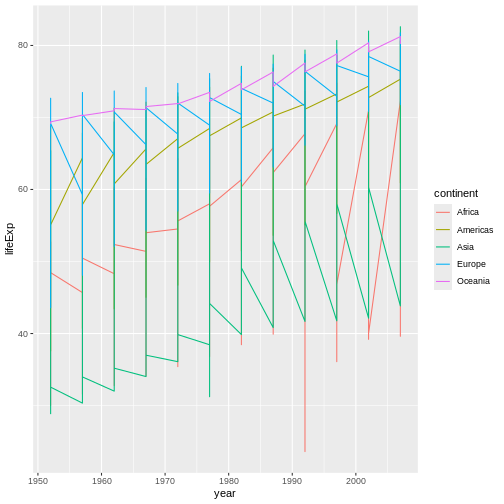
Instead of adding a geom_point layer, we’ve added a
geom_line layer.
However, the result doesn’t look quite as we might have expected: it seems to be jumping around a lot in each continent. Let’s try to separate the data by country, plotting one line for each country:
R
ggplot(data = gapminder, mapping = aes(x=year, y=lifeExp, group=country, color=continent)) +
geom_line()
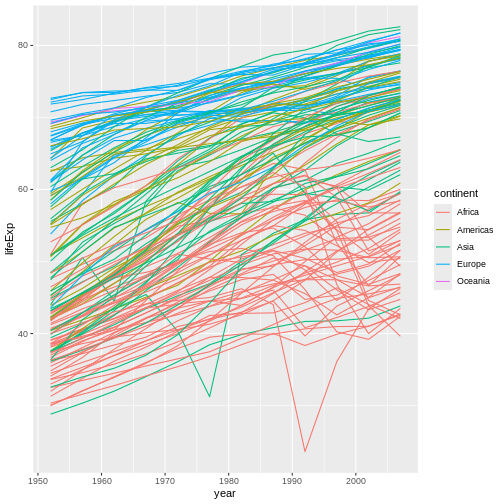
We’ve added the group aesthetic, which
tells ggplot to draw a line for each country.
But what if we want to visualize both lines and points on the plot? We can add another layer to the plot:
R
ggplot(data = gapminder, mapping = aes(x=year, y=lifeExp, group=country, color=continent)) +
geom_line() + geom_point()
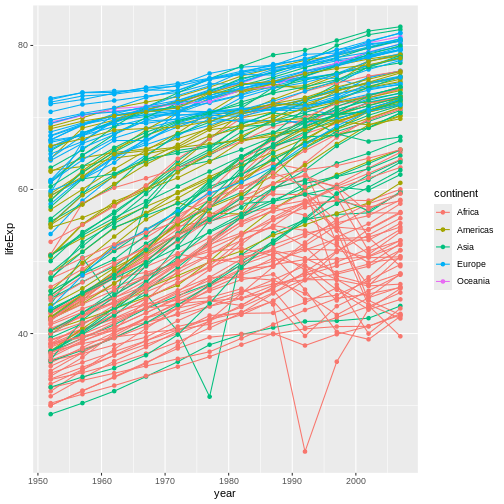
It’s important to note that each layer is drawn on top of the previous layer. In this example, the points have been drawn on top of the lines. Here’s a demonstration:
R
ggplot(data = gapminder, mapping = aes(x=year, y=lifeExp, group=country)) +
geom_line(mapping = aes(color=continent)) + geom_point()
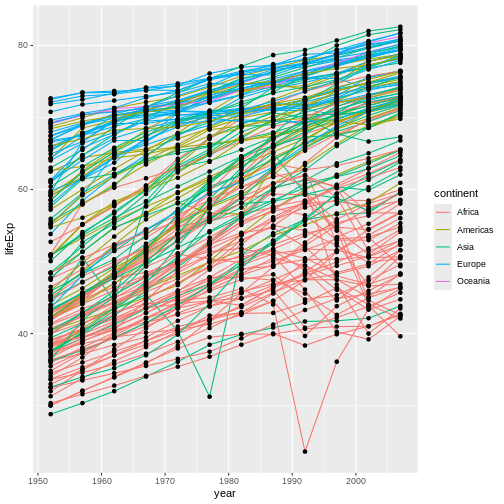
In this example, the aesthetic mapping of
color has been moved from the global plot options in
ggplot to the geom_line layer so it no longer
applies to the points. Now we can clearly see that the points are drawn
on top of the lines.
Tip: Setting an aesthetic to a value instead of a mapping
So far, we’ve seen how to use an aesthetic (such as
color) as a mapping to a variable in the data.
For example, when we use
geom_line(mapping = aes(color=continent)), ggplot will give
a different color to each continent. But what if we want to change the
color of all lines to blue? You may think that
geom_line(mapping = aes(color="blue")) should work, but it
doesn’t. Since we don’t want to create a mapping to a specific variable,
we can move the color specification outside of the aes()
function, like this: geom_line(color="blue").
Challenge 3
Switch the order of the point and line layers from the previous example. What happened?
The lines now get drawn over the points!
R
ggplot(data = gapminder, mapping = aes(x=year, y=lifeExp, group=country)) +
geom_point() + geom_line(mapping = aes(color=continent))
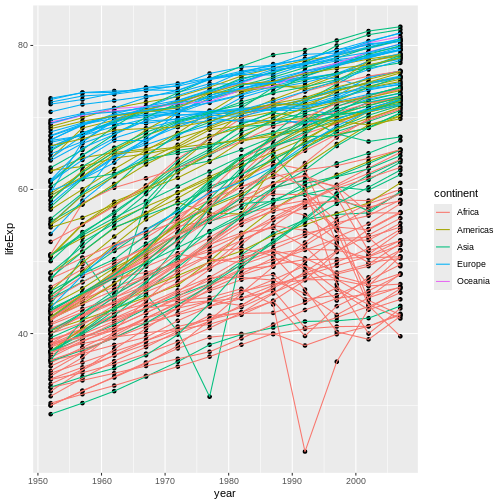
Transformations and statistics
ggplot2 also makes it easy to overlay statistical models over the data. To demonstrate we’ll go back to our first example:
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point()
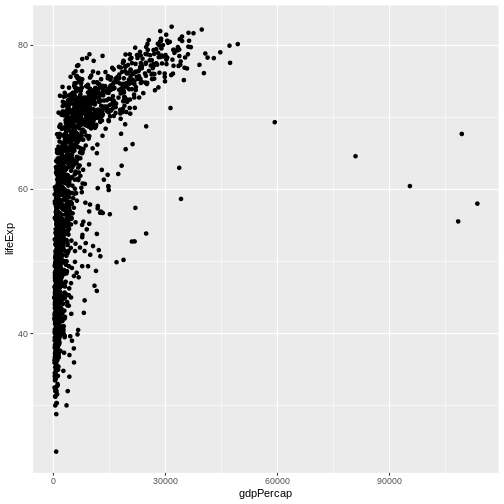
Currently it’s hard to see the relationship between the points due to some strong outliers in GDP per capita. We can change the scale of units on the x axis using the scale functions. These control the mapping between the data values and visual values of an aesthetic. We can also modify the transparency of the points, using the alpha function, which is especially helpful when you have a large amount of data which is very clustered.
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point(alpha = 0.5) + scale_x_log10()
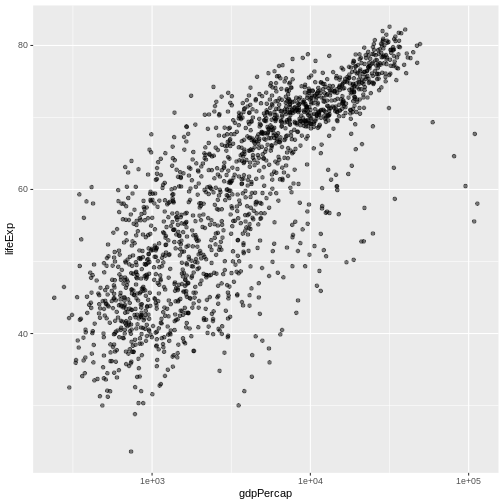
The scale_x_log10 function applied a transformation to
the coordinate system of the plot, so that each multiple of 10 is evenly
spaced from left to right. For example, a GDP per capita of 1,000 is the
same horizontal distance away from a value of 10,000 as the 10,000 value
is from 100,000. This helps to visualize the spread of the data along
the x-axis.
Tip Reminder: Setting an aesthetic to a value instead of a mapping
Notice that we used geom_point(alpha = 0.5). As the
previous tip mentioned, using a setting outside of the
aes() function will cause this value to be used for all
points, which is what we want in this case. But just like any other
aesthetic setting, alpha can also be mapped to a variable in
the data. For example, we can give a different transparency to each
continent with
geom_point(mapping = aes(alpha = continent)).
We can fit a simple relationship to the data by adding another layer,
geom_smooth:
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point(alpha = 0.5) + scale_x_log10() + geom_smooth(method="lm")
OUTPUT
`geom_smooth()` using formula = 'y ~ x'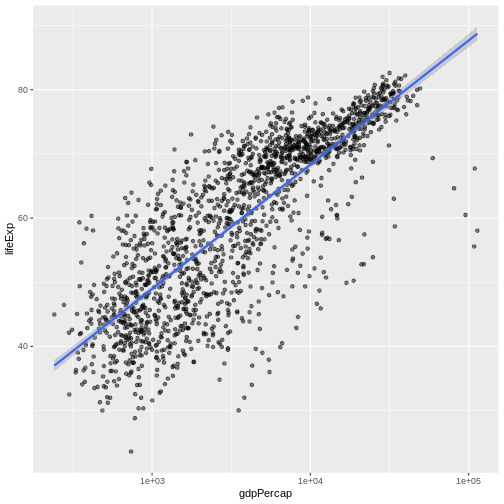
We can make the line thicker by setting the
linewidth aesthetic in the geom_smooth
layer:
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point(alpha = 0.5) + scale_x_log10() + geom_smooth(method="lm", linewidth=1.5)
OUTPUT
`geom_smooth()` using formula = 'y ~ x'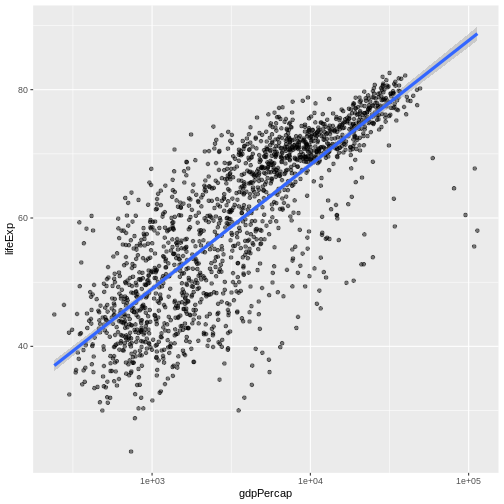
There are two ways an aesthetic can be specified. Here we
set the linewidth aesthetic by passing it as
an argument to geom_smooth and it is applied the same to
the whole geom. Previously in the lesson we’ve used the
aes function to define a mapping between data
variables and their visual representation.
Challenge 4a
Modify the color and size of the points on the point layer in the previous example.
Hint: do not use the aes function.
Hint: the equivalent of linewidth for points is
size.
Here a possible solution: Notice that the color argument
is supplied outside of the aes() function. This means that
it applies to all data points on the graph and is not related to a
specific variable.
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp)) +
geom_point(size=3, color="orange") + scale_x_log10() +
geom_smooth(method="lm", linewidth=1.5)
OUTPUT
`geom_smooth()` using formula = 'y ~ x'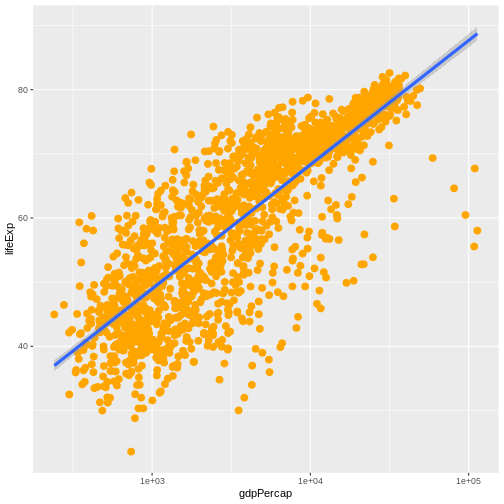
Challenge 4b
Modify your solution to Challenge 4a so that the points are now a different shape and are colored by continent with new trendlines. Hint: The color argument can be used inside the aesthetic.
Here is a possible solution: Notice that supplying the
color argument inside the aes() functions
enables you to connect it to a certain variable. The shape
argument, as you can see, modifies all data points the same way (it is
outside the aes() call) while the color
argument which is placed inside the aes() call modifies a
point’s color based on its continent value.
R
ggplot(data = gapminder, mapping = aes(x = gdpPercap, y = lifeExp, color = continent)) +
geom_point(size=3, shape=17) + scale_x_log10() +
geom_smooth(method="lm", linewidth=1.5)
OUTPUT
`geom_smooth()` using formula = 'y ~ x'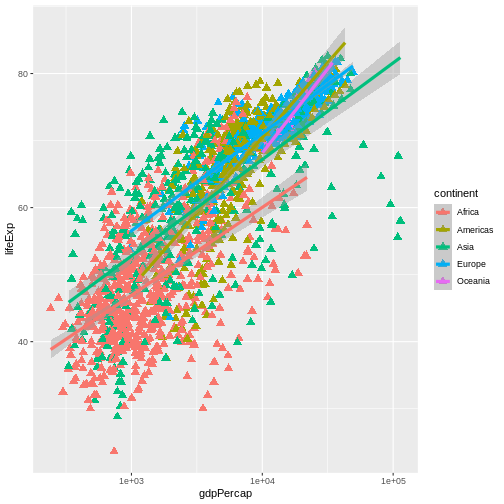
Multi-panel figures
Earlier we visualized the change in life expectancy over time across all countries in one plot. Alternatively, we can split this out over multiple panels by adding a layer of facet panels.
Tip
We start by making a subset of data including only countries located in the Americas. This includes 25 countries, which will begin to clutter the figure. Note that we apply a “theme” definition to rotate the x-axis labels to maintain readability. Nearly everything in ggplot2 is customizable.
R
americas <- gapminder[gapminder$continent == "Americas",]
ggplot(data = americas, mapping = aes(x = year, y = lifeExp)) +
geom_line() +
facet_wrap( ~ country) +
theme(axis.text.x = element_text(angle = 45))
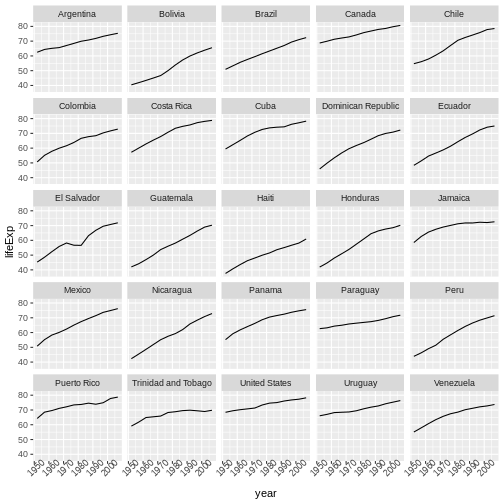
The facet_wrap layer took a “formula” as its argument,
denoted by the tilde (~). This tells R to draw a panel for each unique
value in the country column of the gapminder dataset.
Modifying text
To clean this figure up for a publication we need to change some of the text elements. The x-axis is too cluttered, and the y axis should read “Life expectancy”, rather than the column name in the data frame.
We can do this by adding a couple of different layers. The
theme layer controls the axis text, and overall text
size. Labels for the axes, plot title and any legend can be set using
the labs function. Legend titles are set using the same
names we used in the aes specification. Thus below the
color legend title is set using color = "Continent", while
the title of a fill legend would be set using
fill = "MyTitle".
R
ggplot(data = americas, mapping = aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country) +
labs(
x = "Year", # x axis title
y = "Life expectancy", # y axis title
title = "Figure 1", # main title of figure
color = "Continent" # title of legend
) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
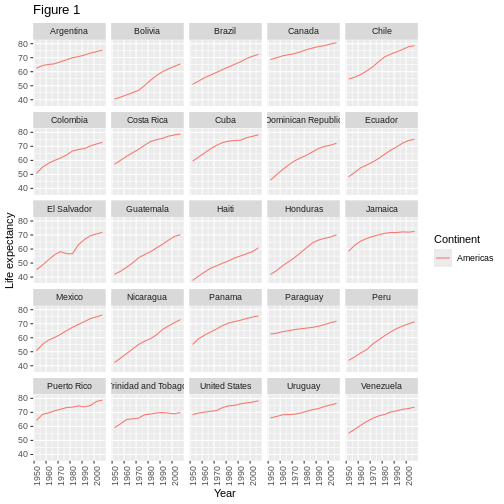
Exporting the plot
The ggsave() function allows you to export a plot
created with ggplot. You can specify the dimension and resolution of
your plot by adjusting the appropriate arguments (width,
height and dpi) to create high quality
graphics for publication. In order to save the plot from above, we first
assign it to a variable lifeExp_plot, then tell
ggsave to save that plot in png format to a
directory called results. (Make sure you have a
results/ folder in your working directory.)
R
lifeExp_plot <- ggplot(data = americas, mapping = aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country) +
labs(
x = "Year", # x axis title
y = "Life expectancy", # y axis title
title = "Figure 1", # main title of figure
color = "Continent" # title of legend
) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
ggsave(filename = "results/lifeExp.png", plot = lifeExp_plot, width = 12, height = 10, dpi = 300, units = "cm")
There are two nice things about ggsave. First, it
defaults to the last plot, so if you omit the plot argument
it will automatically save the last plot you created with
ggplot. Secondly, it tries to determine the format you want
to save your plot in from the file extension you provide for the
filename (for example .png or .pdf). If you
need to, you can specify the format explicitly in the
device argument.
This is a taste of what you can do with ggplot2. RStudio provides a really useful cheat sheet of the different layers available, and more extensive documentation is available on the ggplot2 website. All RStudio cheat sheets are available from the RStudio website. Finally, if you have no idea how to change something, a quick Google search will usually send you to a relevant question and answer on Stack Overflow with reusable code to modify!
Challenge 5
Generate boxplots to compare life expectancy between the different continents during the available years.
Advanced:
- Rename y axis as Life Expectancy.
- Remove x axis labels.
Here a possible solution: xlab() and ylab()
set labels for the x and y axes, respectively The axis title, text and
ticks are attributes of the theme and must be modified within a
theme() call.
R
ggplot(data = gapminder, mapping = aes(x = continent, y = lifeExp, fill = continent)) +
geom_boxplot() + facet_wrap(~year) +
ylab("Life Expectancy") +
theme(axis.title.x=element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
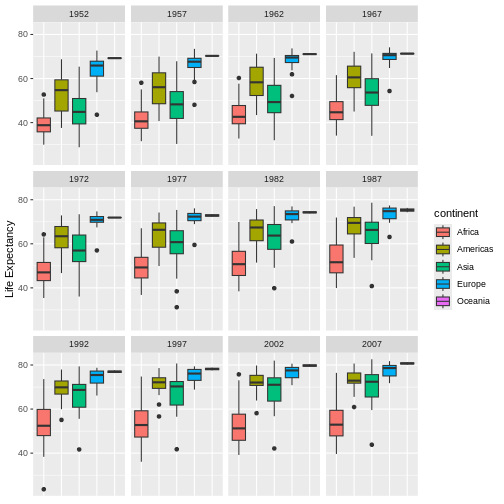
- Use
ggplot2to create plots. - Think about graphics in layers: aesthetics, geometry, statistics, scale transformation, and grouping.
