Overview
Teaching: 30 min Exercises: 15 minQuestions
How can I change the format of dataframes?
Objectives
To be understand the concepts of ‘long’ and ‘wide’ data formats and be able to convert between them with
tidyr.
Researchers often want to manipulate their data from the ‘wide’ to the ‘long’ format, or vice-versa. The ‘long’ format is where:
- each column is a variable
- each row is an observation
In the ‘long’ format, you usually have 1 column for the observed variable and the other columns are ID variables.
For the ‘wide’ format each row is often a site/subject/patient and you have
multiple observation variables containing the same type of data. These can be
either repeated observations over time, or observation of multiple variables (or
a mix of both). You may find data input may be simpler or some other
applications may prefer the ‘wide’ format. However, many of R’s functions have
been designed assuming you have ‘long’ format data. This tutorial will help you
efficiently transform your data regardless of original format.
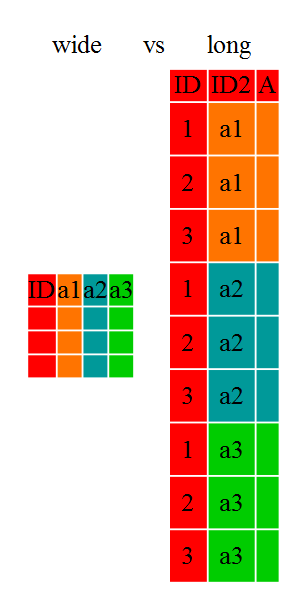
These data formats mainly affect readability. For humans, the wide format is often more intuitive since we can often see more of the data on the screen due to it’s shape. However, the long format is more machine readable and is closer to the formatting of databases. The ID variables in our dataframes are similar to the fields in a database and observed variables are like the database values.
Getting started
First install the packages if you haven’t already done so (you probably installed dplyr in the previous lesson):
#install.packages("tidyr")
#install.packages("dplyr")
Load the packages
library("tidyr")
library("dplyr")
First, lets look at the structure of our original gapminder dataframe:
str(gapminder)
'data.frame': 1704 obs. of 6 variables:
$ country : chr "Afghanistan" "Afghanistan" "Afghanistan" "Afghanistan" ...
$ year : int 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ...
$ pop : num 8425333 9240934 10267083 11537966 13079460 ...
$ continent: chr "Asia" "Asia" "Asia" "Asia" ...
$ lifeExp : num 28.8 30.3 32 34 36.1 ...
$ gdpPercap: num 779 821 853 836 740 ...
Challenge 1
Is gapminder a purely long, purely wide, or some intermediate format?
Solution to Challenge 1
The original gapminder data.frame is in an intermediate format. It is not purely long since it had multiple observation variables (
pop,lifeExp,gdpPercap).
Sometimes, as with the gapminder dataset, we have multiple types of observed
data. It is somewhere in between the purely ‘long’ and ‘wide’ data formats. We
have 3 “ID variables” (continent, country, year) and 3 “Observation
variables” (pop,lifeExp,gdpPercap). I usually prefer my data in this
intermediate format in most cases despite not having ALL observations in 1
column given that all 3 observation variables have different units. There are
few operations that would need us to stretch out this dataframe any longer
(i.e. 4 ID variables and 1 Observation variable).
While using many of the functions in R, which are often vector based, you
usually do not want to do mathematical operations on values with different
units. For example, using the purely long format, a single mean for all of the
values of population, life expectancy, and GDP would not be meaningful since it
would return the mean of values with 3 incompatible units. The solution is that
we first manipulate the data either by grouping (see the lesson on dplyr), or
we change the structure of the dataframe. Note: Some plotting functions in
R actually work better in the wide format data.
From wide to long format with gather()
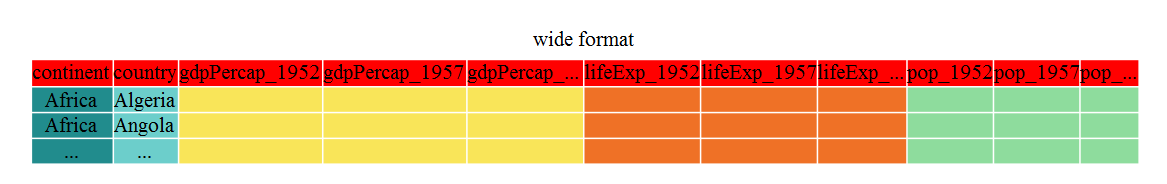
Until now, we’ve been using the nicely formatted original gapminder dataset, but ‘real’ data (i.e. our own research data) will never be so well organized. Here let’s start with the wide format version of the gapminder dataset.
We’ll load the data file and look at it. Note: we don’t want our continent and
country columns to be factors, so we use the stringsAsFactors argument for
read.csv() to disable that.
gap_wide <- read.csv("data/gapminder_wide.csv", stringsAsFactors = FALSE)
str(gap_wide)
'data.frame': 142 obs. of 38 variables:
$ continent : chr "Africa" "Africa" "Africa" "Africa" ...
$ country : chr "Algeria" "Angola" "Benin" "Botswana" ...
$ gdpPercap_1952: num 2449 3521 1063 851 543 ...
$ gdpPercap_1957: num 3014 3828 960 918 617 ...
$ gdpPercap_1962: num 2551 4269 949 984 723 ...
$ gdpPercap_1967: num 3247 5523 1036 1215 795 ...
$ gdpPercap_1972: num 4183 5473 1086 2264 855 ...
$ gdpPercap_1977: num 4910 3009 1029 3215 743 ...
$ gdpPercap_1982: num 5745 2757 1278 4551 807 ...
$ gdpPercap_1987: num 5681 2430 1226 6206 912 ...
$ gdpPercap_1992: num 5023 2628 1191 7954 932 ...
$ gdpPercap_1997: num 4797 2277 1233 8647 946 ...
$ gdpPercap_2002: num 5288 2773 1373 11004 1038 ...
$ gdpPercap_2007: num 6223 4797 1441 12570 1217 ...
$ lifeExp_1952 : num 43.1 30 38.2 47.6 32 ...
$ lifeExp_1957 : num 45.7 32 40.4 49.6 34.9 ...
$ lifeExp_1962 : num 48.3 34 42.6 51.5 37.8 ...
$ lifeExp_1967 : num 51.4 36 44.9 53.3 40.7 ...
$ lifeExp_1972 : num 54.5 37.9 47 56 43.6 ...
$ lifeExp_1977 : num 58 39.5 49.2 59.3 46.1 ...
$ lifeExp_1982 : num 61.4 39.9 50.9 61.5 48.1 ...
$ lifeExp_1987 : num 65.8 39.9 52.3 63.6 49.6 ...
$ lifeExp_1992 : num 67.7 40.6 53.9 62.7 50.3 ...
$ lifeExp_1997 : num 69.2 41 54.8 52.6 50.3 ...
$ lifeExp_2002 : num 71 41 54.4 46.6 50.6 ...
$ lifeExp_2007 : num 72.3 42.7 56.7 50.7 52.3 ...
$ pop_1952 : num 9279525 4232095 1738315 442308 4469979 ...
$ pop_1957 : num 10270856 4561361 1925173 474639 4713416 ...
$ pop_1962 : num 11000948 4826015 2151895 512764 4919632 ...
$ pop_1967 : num 12760499 5247469 2427334 553541 5127935 ...
$ pop_1972 : num 14760787 5894858 2761407 619351 5433886 ...
$ pop_1977 : num 17152804 6162675 3168267 781472 5889574 ...
$ pop_1982 : num 20033753 7016384 3641603 970347 6634596 ...
$ pop_1987 : num 23254956 7874230 4243788 1151184 7586551 ...
$ pop_1992 : num 26298373 8735988 4981671 1342614 8878303 ...
$ pop_1997 : num 29072015 9875024 6066080 1536536 10352843 ...
$ pop_2002 : int 31287142 10866106 7026113 1630347 12251209 7021078 15929988 4048013 8835739 614382 ...
$ pop_2007 : int 33333216 12420476 8078314 1639131 14326203 8390505 17696293 4369038 10238807 710960 ...
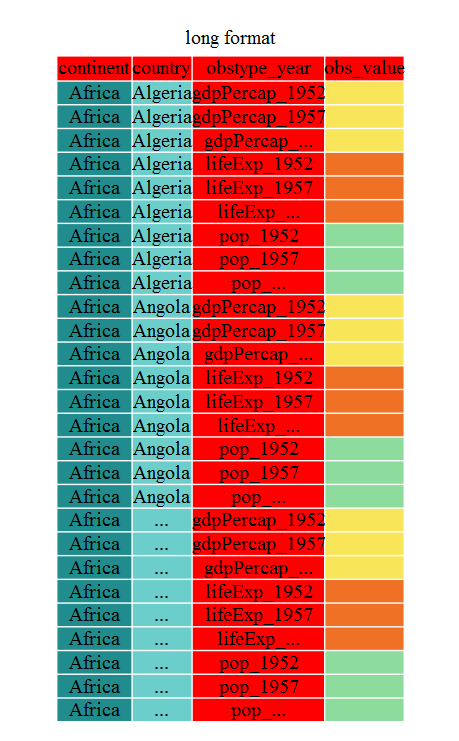
The first step towards getting our nice intermediate data format is to first
convert from the wide to the long format. The tidyr function gather() will
‘gather’ your observation variables into a single variable.
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, starts_with('pop'),
starts_with('lifeExp'), starts_with('gdpPercap'))
str(gap_long)
'data.frame': 5112 obs. of 4 variables:
$ continent : chr "Africa" "Africa" "Africa" "Africa" ...
$ country : chr "Algeria" "Angola" "Benin" "Botswana" ...
$ obstype_year: chr "pop_1952" "pop_1952" "pop_1952" "pop_1952" ...
$ obs_values : num 9279525 4232095 1738315 442308 4469979 ...
Here we have used piping syntax which is similar to what we were doing in the previous lesson with dplyr. In fact, these are compatible and you can use a mix of tidyr and dplyr functions by piping them together
Inside gather() we first name the new column for the new ID variable
(obstype_year), the name for the new amalgamated observation variable
(obs_value), then the names of the old observation variable. We could have
typed out all the observation variables, but as in the select() function (see
dplyr lesson), we can use the starts_with() argument to select all variables
that starts with the desired character string. Gather also allows the alternative
syntax of using the - symbol to identify which variables are not to be
gathered (i.e. ID variables)
gap_long <- gap_wide %>% gather(obstype_year,obs_values,-continent,-country)
str(gap_long)
'data.frame': 5112 obs. of 4 variables:
$ continent : chr "Africa" "Africa" "Africa" "Africa" ...
$ country : chr "Algeria" "Angola" "Benin" "Botswana" ...
$ obstype_year: chr "gdpPercap_1952" "gdpPercap_1952" "gdpPercap_1952" "gdpPercap_1952" ...
$ obs_values : num 2449 3521 1063 851 543 ...
That may seem trivial with this particular dataframe, but sometimes you have 1 ID variable and 40 Observation variables with irregular variables names. The flexibility is a huge time saver!
Now obstype_year actually contains 2 pieces of information, the observation
type (pop,lifeExp, or gdpPercap) and the year. We can use the
separate() function to split the character strings into multiple variables
gap_long <- gap_long %>% separate(obstype_year,into=c('obs_type','year'),sep="_")
gap_long$year <- as.integer(gap_long$year)
Challenge 2
Using
gap_long, calculate the mean life expectancy, population, and gdpPercap for each continent. Hint: use thegroup_by()andsummarize()functions we learned in thedplyrlessonSolution to Challenge 2
gap_long %>% group_by(continent,obs_type) %>% summarize(means=mean(obs_values))Source: local data frame [15 x 3] Groups: continent [?] continent obs_type means <chr> <chr> <dbl> 1 Africa gdpPercap 2.193755e+03 2 Africa lifeExp 4.886533e+01 3 Africa pop 9.916003e+06 4 Americas gdpPercap 7.136110e+03 5 Americas lifeExp 6.465874e+01 6 Americas pop 2.450479e+07 7 Asia gdpPercap 7.902150e+03 8 Asia lifeExp 6.006490e+01 9 Asia pop 7.703872e+07 10 Europe gdpPercap 1.446948e+04 11 Europe lifeExp 7.190369e+01 12 Europe pop 1.716976e+07 13 Oceania gdpPercap 1.862161e+04 14 Oceania lifeExp 7.432621e+01 15 Oceania pop 8.874672e+06
From long to intermediate format with spread()
It is always good to check work. So, let’s use the opposite of gather() to
spread our observation variables back out with the aptly named spread(). We
can then spread our gap_long() to the original intermediate format or the
widest format. Let’s start with the intermediate format.
gap_normal <- gap_long %>% spread(obs_type,obs_values)
dim(gap_normal)
[1] 1704 6
dim(gapminder)
[1] 1704 6
names(gap_normal)
[1] "continent" "country" "year" "gdpPercap" "lifeExp" "pop"
names(gapminder)
[1] "country" "year" "pop" "continent" "lifeExp" "gdpPercap"
Now we’ve got an intermediate dataframe gap_normal with the same dimensions as
the original gapminder, but the order of the variables is different. Let’s fix
that before checking if they are all.equal().
gap_normal <- gap_normal[,names(gapminder)]
all.equal(gap_normal,gapminder)
[1] "Component \"country\": 1704 string mismatches"
[2] "Component \"pop\": Mean relative difference: 1.634504"
[3] "Component \"continent\": 1212 string mismatches"
[4] "Component \"lifeExp\": Mean relative difference: 0.203822"
[5] "Component \"gdpPercap\": Mean relative difference: 1.162302"
head(gap_normal)
country year pop continent lifeExp gdpPercap
1 Algeria 1952 9279525 Africa 43.077 2449.008
2 Algeria 1957 10270856 Africa 45.685 3013.976
3 Algeria 1962 11000948 Africa 48.303 2550.817
4 Algeria 1967 12760499 Africa 51.407 3246.992
5 Algeria 1972 14760787 Africa 54.518 4182.664
6 Algeria 1977 17152804 Africa 58.014 4910.417
head(gapminder)
country year pop continent lifeExp gdpPercap
1 Afghanistan 1952 8425333 Asia 28.801 779.4453
2 Afghanistan 1957 9240934 Asia 30.332 820.8530
3 Afghanistan 1962 10267083 Asia 31.997 853.1007
4 Afghanistan 1967 11537966 Asia 34.020 836.1971
5 Afghanistan 1972 13079460 Asia 36.088 739.9811
6 Afghanistan 1977 14880372 Asia 38.438 786.1134
We’re almost there, the original was sorted by country, continent, then
year.
gap_normal <- gap_normal %>% arrange(country,continent,year)
all.equal(gap_normal,gapminder)
[1] TRUE
That’s great! We’ve gone from the longest format back to the intermediate and we didn’t introduce any errors in our code.
Now lets convert the long all the way back to the wide. In the wide format, we
will keep country and continent as ID variables and spread the observations
across the 3 metrics (pop,lifeExp,gdpPercap) and time (year). First we
need to create appropriate labels for all our new variables (time*metric
combinations) and we also need to unify our ID variables to simplify the process
of defining gap_wide
gap_temp <- gap_long %>% unite(var_ID,continent,country,sep="_")
str(gap_temp)
'data.frame': 5112 obs. of 4 variables:
$ var_ID : chr "Africa_Algeria" "Africa_Angola" "Africa_Benin" "Africa_Botswana" ...
$ obs_type : chr "gdpPercap" "gdpPercap" "gdpPercap" "gdpPercap" ...
$ year : int 1952 1952 1952 1952 1952 1952 1952 1952 1952 1952 ...
$ obs_values: num 2449 3521 1063 851 543 ...
gap_temp <- gap_long %>%
unite(ID_var,continent,country,sep="_") %>%
unite(var_names,obs_type,year,sep="_")
str(gap_temp)
'data.frame': 5112 obs. of 3 variables:
$ ID_var : chr "Africa_Algeria" "Africa_Angola" "Africa_Benin" "Africa_Botswana" ...
$ var_names : chr "gdpPercap_1952" "gdpPercap_1952" "gdpPercap_1952" "gdpPercap_1952" ...
$ obs_values: num 2449 3521 1063 851 543 ...
Using unite() we now have a single ID variable which is a combination of
continent,country,and we have defined variable names. We’re now ready to
pipe in spread()
gap_wide_new <- gap_long %>%
unite(ID_var,continent,country,sep="_") %>%
unite(var_names,obs_type,year,sep="_") %>%
spread(var_names,obs_values)
str(gap_wide_new)
'data.frame': 142 obs. of 37 variables:
$ ID_var : chr "Africa_Algeria" "Africa_Angola" "Africa_Benin" "Africa_Botswana" ...
$ gdpPercap_1952: num 2449 3521 1063 851 543 ...
$ gdpPercap_1957: num 3014 3828 960 918 617 ...
$ gdpPercap_1962: num 2551 4269 949 984 723 ...
$ gdpPercap_1967: num 3247 5523 1036 1215 795 ...
$ gdpPercap_1972: num 4183 5473 1086 2264 855 ...
$ gdpPercap_1977: num 4910 3009 1029 3215 743 ...
$ gdpPercap_1982: num 5745 2757 1278 4551 807 ...
$ gdpPercap_1987: num 5681 2430 1226 6206 912 ...
$ gdpPercap_1992: num 5023 2628 1191 7954 932 ...
$ gdpPercap_1997: num 4797 2277 1233 8647 946 ...
$ gdpPercap_2002: num 5288 2773 1373 11004 1038 ...
$ gdpPercap_2007: num 6223 4797 1441 12570 1217 ...
$ lifeExp_1952 : num 43.1 30 38.2 47.6 32 ...
$ lifeExp_1957 : num 45.7 32 40.4 49.6 34.9 ...
$ lifeExp_1962 : num 48.3 34 42.6 51.5 37.8 ...
$ lifeExp_1967 : num 51.4 36 44.9 53.3 40.7 ...
$ lifeExp_1972 : num 54.5 37.9 47 56 43.6 ...
$ lifeExp_1977 : num 58 39.5 49.2 59.3 46.1 ...
$ lifeExp_1982 : num 61.4 39.9 50.9 61.5 48.1 ...
$ lifeExp_1987 : num 65.8 39.9 52.3 63.6 49.6 ...
$ lifeExp_1992 : num 67.7 40.6 53.9 62.7 50.3 ...
$ lifeExp_1997 : num 69.2 41 54.8 52.6 50.3 ...
$ lifeExp_2002 : num 71 41 54.4 46.6 50.6 ...
$ lifeExp_2007 : num 72.3 42.7 56.7 50.7 52.3 ...
$ pop_1952 : num 9279525 4232095 1738315 442308 4469979 ...
$ pop_1957 : num 10270856 4561361 1925173 474639 4713416 ...
$ pop_1962 : num 11000948 4826015 2151895 512764 4919632 ...
$ pop_1967 : num 12760499 5247469 2427334 553541 5127935 ...
$ pop_1972 : num 14760787 5894858 2761407 619351 5433886 ...
$ pop_1977 : num 17152804 6162675 3168267 781472 5889574 ...
$ pop_1982 : num 20033753 7016384 3641603 970347 6634596 ...
$ pop_1987 : num 23254956 7874230 4243788 1151184 7586551 ...
$ pop_1992 : num 26298373 8735988 4981671 1342614 8878303 ...
$ pop_1997 : num 29072015 9875024 6066080 1536536 10352843 ...
$ pop_2002 : num 31287142 10866106 7026113 1630347 12251209 ...
$ pop_2007 : num 33333216 12420476 8078314 1639131 14326203 ...
Challenge 3
Take this 1 step further and create a
gap_ludicrously_wideformat data by spreading over countries, year and the 3 metrics? Hint this new dataframe should only have 5 rows.Solution to Challenge 3
gap_ludicrously_wide <- gap_long %>% unite(var_names,obs_type,year,country,sep="_") %>% spread(var_names,obs_values)
Now we have a great ‘wide’ format dataframe, but the ID_var could be more
usable, let’s separate it into 2 variables with separate()
gap_wide_betterID <- separate(gap_wide_new,ID_var,c("continent","country"),sep="_")
gap_wide_betterID <- gap_long %>%
unite(ID_var, continent,country,sep="_") %>%
unite(var_names, obs_type,year,sep="_") %>%
spread(var_names, obs_values) %>%
separate(ID_var, c("continent","country"),sep="_")
str(gap_wide_betterID)
'data.frame': 142 obs. of 38 variables:
$ continent : chr "Africa" "Africa" "Africa" "Africa" ...
$ country : chr "Algeria" "Angola" "Benin" "Botswana" ...
$ gdpPercap_1952: num 2449 3521 1063 851 543 ...
$ gdpPercap_1957: num 3014 3828 960 918 617 ...
$ gdpPercap_1962: num 2551 4269 949 984 723 ...
$ gdpPercap_1967: num 3247 5523 1036 1215 795 ...
$ gdpPercap_1972: num 4183 5473 1086 2264 855 ...
$ gdpPercap_1977: num 4910 3009 1029 3215 743 ...
$ gdpPercap_1982: num 5745 2757 1278 4551 807 ...
$ gdpPercap_1987: num 5681 2430 1226 6206 912 ...
$ gdpPercap_1992: num 5023 2628 1191 7954 932 ...
$ gdpPercap_1997: num 4797 2277 1233 8647 946 ...
$ gdpPercap_2002: num 5288 2773 1373 11004 1038 ...
$ gdpPercap_2007: num 6223 4797 1441 12570 1217 ...
$ lifeExp_1952 : num 43.1 30 38.2 47.6 32 ...
$ lifeExp_1957 : num 45.7 32 40.4 49.6 34.9 ...
$ lifeExp_1962 : num 48.3 34 42.6 51.5 37.8 ...
$ lifeExp_1967 : num 51.4 36 44.9 53.3 40.7 ...
$ lifeExp_1972 : num 54.5 37.9 47 56 43.6 ...
$ lifeExp_1977 : num 58 39.5 49.2 59.3 46.1 ...
$ lifeExp_1982 : num 61.4 39.9 50.9 61.5 48.1 ...
$ lifeExp_1987 : num 65.8 39.9 52.3 63.6 49.6 ...
$ lifeExp_1992 : num 67.7 40.6 53.9 62.7 50.3 ...
$ lifeExp_1997 : num 69.2 41 54.8 52.6 50.3 ...
$ lifeExp_2002 : num 71 41 54.4 46.6 50.6 ...
$ lifeExp_2007 : num 72.3 42.7 56.7 50.7 52.3 ...
$ pop_1952 : num 9279525 4232095 1738315 442308 4469979 ...
$ pop_1957 : num 10270856 4561361 1925173 474639 4713416 ...
$ pop_1962 : num 11000948 4826015 2151895 512764 4919632 ...
$ pop_1967 : num 12760499 5247469 2427334 553541 5127935 ...
$ pop_1972 : num 14760787 5894858 2761407 619351 5433886 ...
$ pop_1977 : num 17152804 6162675 3168267 781472 5889574 ...
$ pop_1982 : num 20033753 7016384 3641603 970347 6634596 ...
$ pop_1987 : num 23254956 7874230 4243788 1151184 7586551 ...
$ pop_1992 : num 26298373 8735988 4981671 1342614 8878303 ...
$ pop_1997 : num 29072015 9875024 6066080 1536536 10352843 ...
$ pop_2002 : num 31287142 10866106 7026113 1630347 12251209 ...
$ pop_2007 : num 33333216 12420476 8078314 1639131 14326203 ...
all.equal(gap_wide, gap_wide_betterID)
[1] TRUE
There and back again!
Other great resources
Key Points
Use the
tidyrpackage to change the layout of dataframes.Use
gather()to go from wide to long format.Use
scatter()to go from long to wide format.