Content from Introduction
Last updated on 2023-04-25 | Edit this page
Estimated time: 25 minutes
Overview
Questions
- How can I make my results easier to reproduce?
Objectives
- Explain what Make is for.
- Explain why Make differs from shell scripts.
- Name other popular build tools.
Let’s imagine that we’re interested in testing Zipf’s Law in some of our favorite books.
Zipf’s Law
The most frequently-occurring word occurs approximately twice as often as the second most frequent word. This is Zipf’s Law.
We’ve compiled our raw data i.e. the books we want to analyze and have prepared several Python scripts that together make up our analysis pipeline.
Let’s take quick look at one of the books using the command
head books/isles.txt.
Our directory has the Python scripts and data files we will be working with:
OUTPUT
|- books
| |- abyss.txt
| |- isles.txt
| |- last.txt
| |- LICENSE_TEXTS.md
| |- sierra.txt
|- plotcounts.py
|- countwords.py
|- testzipf.pyThe first step is to count the frequency of each word in a book. For
this purpose we will use a python script countwords.py
which takes two command line arguments. The first argument is the input
file (books/isles.txt) and the second is the output file
that is generated (here isles.dat) by processing the
input.
Let’s take a quick peek at the result.
This shows us the top 5 lines in the output file:
OUTPUT
the 3822 6.7371760973
of 2460 4.33632998414
and 1723 3.03719372466
to 1479 2.60708619778
a 1308 2.30565838181We can see that the file consists of one row per word. Each row shows the word itself, the number of occurrences of that word, and the number of occurrences as a percentage of the total number of words in the text file.
We can do the same thing for a different book:
OUTPUT
the 4044 6.35449402891
and 2807 4.41074795726
of 1907 2.99654305468
a 1594 2.50471401634
to 1515 2.38057825267Let’s visualize the results. The script plotcounts.py
reads in a data file and plots the 10 most frequently occurring words as
a text-based bar plot:
OUTPUT
the ########################################################################
of ##############################################
and ################################
to ############################
a #########################
in ###################
is #################
that ############
by ###########
it ###########plotcounts.py can also show the plot graphically:
Close the window to exit the plot.
plotcounts.py can also create the plot as an image file
(e.g. a PNG file):
Finally, let’s test Zipf’s law for these books:
OUTPUT
Book First Second Ratio
abyss 4044 2807 1.44
isles 3822 2460 1.55So we’re not too far off from Zipf’s law.
Together these scripts implement a common workflow:
- Read a data file.
- Perform an analysis on this data file.
- Write the analysis results to a new file.
- Plot a graph of the analysis results.
- Save the graph as an image, so we can put it in a paper.
- Make a summary table of the analyses
Running countwords.py and plotcounts.py at
the shell prompt, as we have been doing, is fine for one or two files.
If, however, we had 5 or 10 or 20 text files, or if the number of steps
in the pipeline were to expand, this could turn into a lot of work.
Plus, no one wants to sit and wait for a command to finish, even just
for 30 seconds.
The most common solution to the tedium of data processing is to write a shell script that runs the whole pipeline from start to finish.
So to reproduce the tasks that we have just done we create a new file
named run_pipeline.sh in which we place the commands one by
one. Using a text editor of your choice (e.g. for nano use the command
nano run_pipeline.sh) copy and paste the following text and
save it.
BASH
# USAGE: bash run_pipeline.sh
# to produce plots for isles and abyss
# and the summary table for the Zipf's law tests
python countwords.py books/isles.txt isles.dat
python countwords.py books/abyss.txt abyss.dat
python plotcounts.py isles.dat isles.png
python plotcounts.py abyss.dat abyss.png
# Generate summary table
python testzipf.py abyss.dat isles.dat > results.txtRun the script and check that the output is the same as before:
This shell script solves several problems in computational reproducibility:
- It explicitly documents our pipeline, making communication with colleagues (and our future selves) more efficient.
- It allows us to type a single command,
bash run_pipeline.sh, to reproduce the full analysis. - It prevents us from repeating typos or mistakes. You might not get it right the first time, but once you fix something it’ll stay fixed.
Despite these benefits it has a few shortcomings.
Let’s adjust the width of the bars in our plot produced by
plotcounts.py.
Edit plotcounts.py so that the bars are 0.8 units wide
instead of 1 unit. (Hint: replace width = 1.0 with
width = 0.8 in the definition of
plot_word_counts.)
Now we want to recreate our figures. We could just
bash run_pipeline.sh again. That would work, but it could
also be a big pain if counting words takes more than a few seconds. The
word counting routine hasn’t changed; we shouldn’t need to recreate
those files.
Alternatively, we could manually rerun the plotting for each word-count file. (Experienced shell scripters can make this easier on themselves using a for-loop.)
With this approach, however, we don’t get many of the benefits of having a shell script in the first place.
Another popular option is to comment out a subset of the lines in
run_pipeline.sh:
BASH
# USAGE: bash run_pipeline.sh
# to produce plots for isles and abyss
# and the summary table for the Zipf's law tests.
# These lines are commented out because they don't need to be rerun.
#python countwords.py books/isles.txt isles.dat
#python countwords.py books/abyss.txt abyss.dat
python plotcounts.py isles.dat isles.png
python plotcounts.py abyss.dat abyss.png
# Generate summary table
# This line is also commented out because it doesn't need to be rerun.
#python testzipf.py abyss.dat isles.dat > results.txtThen, we would run our modified shell script using
bash run_pipeline.sh.
But commenting out these lines, and subsequently uncommenting them, can be a hassle and source of errors in complicated pipelines.
What we really want is an executable description of our pipeline that allows software to do the tricky part for us: figuring out what steps need to be rerun.
For our pipeline Make can execute the commands needed to run our analysis and plot our results. Like shell scripts it allows us to execute complex sequences of commands via a single shell command. Unlike shell scripts it explicitly records the dependencies between files - what files are needed to create what other files - and so can determine when to recreate our data files or image files, if our text files change. Make can be used for any commands that follow the general pattern of processing files to create new files, for example:
- Run analysis scripts on raw data files to get data files that summarize the raw data (e.g. creating files with word counts from book text).
- Run visualization scripts on data files to produce plots (e.g. creating images of word counts).
- Parse and combine text files and plots to create papers.
- Compile source code into executable programs or libraries.
There are now many build tools available, for example Apache ANT, doit, and nmake for Windows. Which is best for you depends on your requirements, intended usage, and operating system. However, they all share the same fundamental concepts as Make.
Also, you might come across build generation scripts e.g. GNU Autoconf and CMake. Those tools do not run the pipelines directly, but rather generate files for use with the build tools.
Why Use Make if it is Almost 40 Years Old?
Make development was started by Stuart Feldman in 1977 as a Bell Labs summer intern. Since then it has been undergoing an active development and several implementations are available. Since it solves a common issue of workflow management, it remains in widespread use even today.
Researchers working with legacy codes in C or FORTRAN, which are very common in high-performance computing, will, very likely encounter Make.
Researchers can use Make for implementing reproducible research workflows, automating data analysis and visualisation (using Python or R) and combining tables and plots with text to produce reports and papers for publication.
Make’s fundamental concepts are common across build tools.
GNU Make is a free-libre, fast, well-documented, and very popular Make implementation. From now on, we will focus on it, and when we say Make, we mean GNU Make.
- Make allows us to specify what depends on what and how to update things that are out of date.
Content from Makefiles
Last updated on 2023-04-24 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- How do I write a simple Makefile?
Objectives
- Recognize the key parts of a Makefile, rules, targets, dependencies and actions.
- Write a simple Makefile.
- Run Make from the shell.
- Explain when and why to mark targets as
.PHONY. - Explain constraints on dependencies.
Create a file, called Makefile, with the following
content:
# Count words.
isles.dat : books/isles.txt
python countwords.py books/isles.txt isles.datThis is a build file, which for Make is called a Makefile - a file executed by Make. Note how it resembles one of the lines from our shell script.
Let us go through each line in turn:
-
#denotes a comment. Any text from#to the end of the line is ignored by Make but could be very helpful for anyone reading your Makefile. -
isles.datis a target, a file to be created, or built. -
books/isles.txtis a dependency, a file that is needed to build or update the target. Targets can have zero or more dependencies. - A colon,
:, separates targets from dependencies. -
python countwords.py books/isles.txt isles.datis an action, a command to run to build or update the target using the dependencies. Targets can have zero or more actions. These actions form a recipe to build the target from its dependencies and are executed similarly to a shell script. - Actions are indented using a single TAB character, not 8 spaces. This is a legacy of Make’s 1970’s origins. If the difference between spaces and a TAB character isn’t obvious in your editor, try moving your cursor from one side of the TAB to the other. It should jump four or more spaces.
- Together, the target, dependencies, and actions form a rule.
Our rule above describes how to build the target
isles.dat using the action
python countwords.py and the dependency
books/isles.txt.
Information that was implicit in our shell script - that we are
generating a file called isles.dat and that creating this
file requires books/isles.txt - is now made explicit by
Make’s syntax.
Let’s first ensure we start from scratch and delete the
.dat and .png files we created earlier:
By default, Make looks for a Makefile, called Makefile,
and we can run Make as follows:
By default, Make prints out the actions it executes:
OUTPUT
python countwords.py books/isles.txt isles.datIf we see,
ERROR
Makefile:3: *** missing separator. Stop.then we have used a space instead of a TAB characters to indent one of our actions.
Let’s see if we got what we expected.
The first 5 lines of isles.dat should look exactly like
before.
Makefiles Do Not Have to be Called
Makefile
We don’t have to call our Makefile Makefile. However, if
we call it something else we need to tell Make where to find it. This we
can do using -f flag. For example, if our Makefile is named
MyOtherMakefile:
Sometimes, the suffix .mk will be used to identify
Makefiles that are not called Makefile
e.g. install.mk, common.mk etc.
When we re-run our Makefile, Make now informs us that:
OUTPUT
make: `isles.dat' is up to date.This is because our target, isles.dat, has now been
created, and Make will not create it again. To see how this works, let’s
pretend to update one of the text files. Rather than opening the file in
an editor, we can use the shell touch command to update its
timestamp (which would happen if we did edit the file):
If we compare the timestamps of books/isles.txt and
isles.dat,
then we see that isles.dat, the target, is now older
than books/isles.txt, its dependency:
OUTPUT
-rw-r--r-- 1 mjj Administ 323972 Jun 12 10:35 books/isles.txt
-rw-r--r-- 1 mjj Administ 182273 Jun 12 09:58 isles.datIf we run Make again,
then it recreates isles.dat:
OUTPUT
python countwords.py books/isles.txt isles.datWhen it is asked to build a target, Make checks the ‘last modification time’ of both the target and its dependencies. If any dependency has been updated since the target, then the actions are re-run to update the target. Using this approach, Make knows to only rebuild the files that, either directly or indirectly, depend on the file that changed. This is called an incremental build.
Makefiles as Documentation
By explicitly recording the inputs to and outputs from steps in our analysis and the dependencies between files, Makefiles act as a type of documentation, reducing the number of things we have to remember.
Let’s add another rule to the end of Makefile:
abyss.dat : books/abyss.txt
python countwords.py books/abyss.txt abyss.datIf we run Make,
then we get:
OUTPUT
make: `isles.dat' is up to date.Nothing happens because Make attempts to build the first target it
finds in the Makefile, the default target, which is
isles.dat which is already up-to-date. We need to
explicitly tell Make we want to build abyss.dat:
Now, we get:
OUTPUT
python countwords.py books/abyss.txt abyss.dat“Up to Date” Versus “Nothing to be Done”
If we ask Make to build a file that already exists and is up to date, then Make informs us that:
OUTPUT
make: `isles.dat' is up to date.If we ask Make to build a file that exists but for which there is no rule in our Makefile, then we get message like:
OUTPUT
make: Nothing to be done for `countwords.py'.up to date means that the Makefile has a rule with one
or more actions whose target is the name of a file (or directory) and
the file is up to date.
Nothing to be done means that the file exists but either
:
- the Makefile has no rule for it, or
- the Makefile has a rule for it, but that rule has no actions
We may want to remove all our data files so we can explicitly
recreate them all. We can introduce a new target, and associated rule,
to do this. We will call it clean, as this is a common name
for rules that delete auto-generated files, like our .dat
files:
clean :
rm -f *.datThis is an example of a rule that has no dependencies.
clean has no dependencies on any .dat file as
it makes no sense to create these just to remove them. We just want to
remove the data files whether or not they exist. If we run Make and
specify this target,
then we get:
OUTPUT
rm -f *.datThere is no actual thing built called clean. Rather, it
is a short-hand that we can use to execute a useful sequence of actions.
Such targets, though very useful, can lead to problems. For example, let
us recreate our data files, create a directory called
clean, then run Make:
We get:
OUTPUT
make: `clean' is up to date.Make finds a file (or directory) called clean and, as
its clean rule has no dependencies, assumes that
clean has been built and is up-to-date and so does not
execute the rule’s actions. As we are using clean as a
short-hand, we need to tell Make to always execute this rule if we run
make clean, by telling Make that this is a phony target, that it does not
build anything. This we do by marking the target as
.PHONY:
.PHONY : clean
clean :
rm -f *.datIf we run Make,
then we get:
OUTPUT
rm -f *.datWe can add a similar command to create all the data files. We can put
this at the top of our Makefile so that it is the default target, which is
executed by default if no target is given to the make
command:
.PHONY : dats
dats : isles.dat abyss.datThis is an example of a rule that has dependencies that are targets of other rules. When Make runs, it will check to see if the dependencies exist and, if not, will see if rules are available that will create these. If such rules exist it will invoke these first, otherwise Make will raise an error.
Dependencies
The order of rebuilding dependencies is arbitrary. You should not assume that they will be built in the order in which they are listed.
Dependencies must form a directed acyclic graph. A target cannot depend on a dependency which itself, or one of its dependencies, depends on that target.
This rule (dats) is also an example of a rule that has
no actions. It is used purely to trigger the build of its dependencies,
if needed.
If we run,
then Make creates the data files:
OUTPUT
python countwords.py books/isles.txt isles.dat
python countwords.py books/abyss.txt abyss.datIf we run make dats again, then Make will see that the
dependencies (isles.dat and abyss.dat) are
already up to date. Given the target dats has no actions,
there is nothing to be done:
OUTPUT
make: Nothing to be done for `dats'.Our Makefile now looks like this:
# Count words.
.PHONY : dats
dats : isles.dat abyss.dat
isles.dat : books/isles.txt
python countwords.py books/isles.txt isles.dat
abyss.dat : books/abyss.txt
python countwords.py books/abyss.txt abyss.dat
.PHONY : clean
clean :
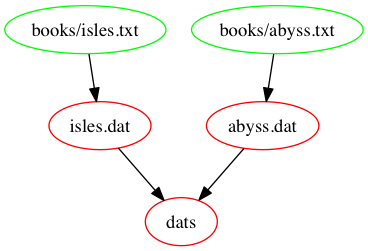
rm -f *.datThe following figure shows a graph of the dependencies embodied
within our Makefile, involved in building the dats
target:
Write Two New Rules
- Write a new rule for
last.dat, created frombooks/last.txt. - Update the
datsrule with this target. - Write a new rule for
results.txt, which creates the summary table. The rule needs to:
- Depend upon each of the three
.datfiles. - Invoke the action
python testzipf.py abyss.dat isles.dat last.dat > results.txt.
- Put this rule at the top of the Makefile so that it is the default target.
- Update
cleanso that it removesresults.txt.
The starting Makefile is here.
See this file for a solution.
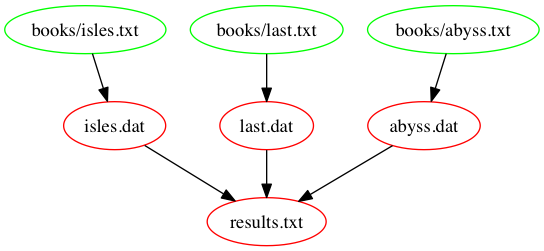
The following figure shows the dependencies embodied within our
Makefile, involved in building the results.txt target:
- Use
#for comments in Makefiles. - Write rules as
target: dependencies. - Specify update actions in a tab-indented block under the rule.
- Use
.PHONYto mark targets that don’t correspond to files.
Content from Automatic Variables
Last updated on 2023-04-24 | Edit this page
Estimated time: 15 minutes
Overview
Questions
- How can I abbreviate the rules in my Makefiles?
Objectives
- Use Make automatic variables to remove duplication in a Makefile.
- Explain why shell wildcards in dependencies can cause problems.
After the exercise at the end of the previous episode, our Makefile looked like this:
# Generate summary table.
results.txt : isles.dat abyss.dat last.dat
python testzipf.py abyss.dat isles.dat last.dat > results.txt
# Count words.
.PHONY : dats
dats : isles.dat abyss.dat last.dat
isles.dat : books/isles.txt
python countwords.py books/isles.txt isles.dat
abyss.dat : books/abyss.txt
python countwords.py books/abyss.txt abyss.dat
last.dat : books/last.txt
python countwords.py books/last.txt last.dat
.PHONY : clean
clean :
rm -f *.dat
rm -f results.txtOur Makefile has a lot of duplication. For example, the names of text files and data files are repeated in many places throughout the Makefile. Makefiles are a form of code and, in any code, repeated code can lead to problems e.g. we rename a data file in one part of the Makefile but forget to rename it elsewhere.
D.R.Y. (Don’t Repeat Yourself)
In many programming languages, the bulk of the language features are there to allow the programmer to describe long-winded computational routines as short, expressive, beautiful code. Features in Python or R or Java, such as user-defined variables and functions are useful in part because they mean we don’t have to write out (or think about) all of the details over and over again. This good habit of writing things out only once is known as the “Don’t Repeat Yourself” principle or D.R.Y.
Let us set about removing some of the repetition from our Makefile.
In our results.txt rule we duplicate the data file names
and the name of the results file name:
results.txt : isles.dat abyss.dat last.dat
python testzipf.py abyss.dat isles.dat last.dat > results.txtLooking at the results file name first, we can replace it in the
action with $@:
results.txt : isles.dat abyss.dat last.dat
python testzipf.py abyss.dat isles.dat last.dat > $@$@ is a Make automatic variable which
means ‘the target of the current rule’. When Make is run it will replace
this variable with the target name.
We can replace the dependencies in the action with
$^:
results.txt : isles.dat abyss.dat last.dat
python testzipf.py $^ > $@$^ is another automatic variable which means ‘all the
dependencies of the current rule’. Again, when Make is run it will
replace this variable with the dependencies.
Let’s update our text files and re-run our rule:
We get:
OUTPUT
python countwords.py books/isles.txt isles.dat
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/last.txt last.dat
python testzipf.py isles.dat abyss.dat last.dat > results.txtAs we saw, $^ means ‘all the dependencies of the current
rule’. This works well for results.txt as its action treats
all the dependencies the same - as the input for the
testzipf.py script.
However, for some rules, we may want to treat the first dependency
differently. For example, our rules for .dat use their
first (and only) dependency specifically as the input file to
countwords.py. If we add additional dependencies (as we
will soon do) then we don’t want these being passed as input files to
countwords.py as it expects only one input file to be named
when it is invoked.
Make provides an automatic variable for this, $<
which means ‘the first dependency of the current rule’.
Rewrite .dat Rules to Use
Automatic Variables
Rewrite each .dat rule to use the automatic variables
$@ (‘the target of the current rule’) and
$< (‘the first dependency of the current rule’). This file contains the
Makefile immediately before the challenge.
See this file for a solution to this challenge.
- Use
$@to refer to the target of the current rule. - Use
$^to refer to the dependencies of the current rule. - Use
$<to refer to the first dependency of the current rule.
Content from Dependencies on Data and Code
Last updated on 2023-04-24 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- How can I write a Makefile to update things when my scripts have changed rather than my input files?
Objectives
- Output files are a product not only of input files but of the scripts or code that created the output files.
- Recognize and avoid false dependencies.
Our Makefile now looks like this:
# Generate summary table.
results.txt : isles.dat abyss.dat last.dat
python testzipf.py $^ > $@
# Count words.
.PHONY : dats
dats : isles.dat abyss.dat last.dat
isles.dat : books/isles.txt
python countwords.py $< $@
abyss.dat : books/abyss.txt
python countwords.py $< $@
last.dat : books/last.txt
python countwords.py $< $@
.PHONY : clean
clean :
rm -f *.dat
rm -f results.txtOur data files are produced using not only the input text files but
also the script countwords.py that processes the text files
and creates the data files. A change to countwords.py
(e.g. adding a new column of summary data or removing an existing one)
results in changes to the .dat files it outputs. So, let’s
pretend to edit countwords.py, using touch,
and re-run Make:
Nothing happens! Though we’ve updated countwords.py our
data files are not updated because our rules for creating
.dat files don’t record any dependencies on
countwords.py.
We need to add countwords.py as a dependency of each of
our data files also:
isles.dat : books/isles.txt countwords.py
python countwords.py $< $@
abyss.dat : books/abyss.txt countwords.py
python countwords.py $< $@
last.dat : books/last.txt countwords.py
python countwords.py $< $@If we pretend to edit countwords.py and re-run Make,
then we get:
OUTPUT
python countwords.py books/isles.txt isles.dat
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/last.txt last.datDry run
make can show the commands it will execute without
actually running them if we pass the -n flag:
This gives the same output to the screen as without the
-n flag, but the commands are not actually run. Using this
‘dry-run’ mode is a good way to check that you have set up your Makefile
properly before actually running the commands in it.
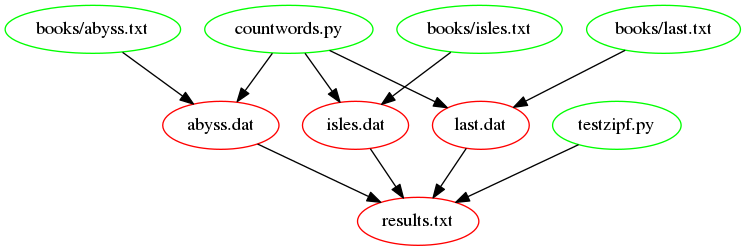
The following figure shows a graph of the dependencies, that are
involved in building the target results.txt. Notice the
recently added dependencies countwords.py and
testzipf.py. This is how the Makefile should look after
completing the rest of the exercises in this episode.
Why Don’t the .txt Files Depend
on countwords.py?
.txt files are input files and as such have no
dependencies. To make these depend on countwords.py would
introduce a false
dependency which is not desirable.
Intuitively, we should also add countwords.py as a
dependency for results.txt, because the final table should
be rebuilt if we remake the .dat files. However, it turns
out we don’t have to do that! Let’s see what happens to
results.txt when we update countwords.py:
then we get:
OUTPUT
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/isles.txt isles.dat
python countwords.py books/last.txt last.dat
python testzipf.py abyss.dat isles.dat last.dat > results.txtThe whole pipeline is triggered, even the creation of the
results.txt file! To understand this, note that according
to the dependency figure, results.txt depends on the
.dat files. The update of countwords.py
triggers an update of the *.dat files. Thus,
make sees that the dependencies (the .dat
files) are newer than the target file (results.txt) and
thus it recreates results.txt. This is an example of the
power of make: updating a subset of the files in the
pipeline triggers rerunning the appropriate downstream steps.
3. only last.dat and
results.txt are recreated.
Follow the dependency tree to understand the answer(s).
testzipf.py as a Dependency of
results.txt.
What would happen if you added testzipf.py as dependency
of results.txt, and why?
If you change the rule for the results.txt file like
this:
results.txt : isles.dat abyss.dat last.dat testzipf.py
python testzipf.py $^ > $@testzipf.py becomes a part of $^, thus the
command becomes
This results in an error from testzipf.py as it tries to
parse the script as if it were a .dat file. Try this by
running:
You’ll get
ERROR
python testzipf.py abyss.dat isles.dat last.dat testzipf.py > results.txt
Traceback (most recent call last):
File "testzipf.py", line 19, in <module>
counts = load_word_counts(input_file)
File "path/to/testzipf.py", line 39, in load_word_counts
counts.append((fields[0], int(fields[1]), float(fields[2])))
IndexError: list index out of range
make: *** [results.txt] Error 1We still have to add the testzipf.py script as
dependency to results.txt. Given the answer to the
challenge above, we need to make a couple of small changes so that we
can still use automatic variables.
We’ll move testzipf.py to be the first dependency and
then edit the action so that we pass all the dependencies as arguments
to python using $^.
results.txt : testzipf.py isles.dat abyss.dat last.dat
python $^ > $@- Make results depend on processing scripts as well as data files.
- Dependencies are transitive: if A depends on B and B depends on C, a change to C will indirectly trigger an update to A.
Content from Pattern Rules
Last updated on 2023-04-24 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- How can I define rules to operate on similar files?
Objectives
- Write Make pattern rules.
Our Makefile still has repeated content. The rules for each
.dat file are identical apart from the text and data file
names. We can replace these rules with a single pattern rule which can be used to
build any .dat file from a .txt file in
books/:
%.dat : countwords.py books/%.txt
python $^ $@% is a Make wildcard, matching any number of any
characters.
This rule can be interpreted as: “In order to build a file named
[something].dat (the target) find a file named
books/[that same something].txt (one of the dependencies)
and run python [the dependencies] [the target].”
If we re-run Make,
then we get:
OUTPUT
python countwords.py books/isles.txt isles.dat
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/last.txt last.datNote that we can still use Make to build individual .dat
targets as before, and that our new rule will work no matter what stem
is being matched.
which gives the output below:
OUTPUT
python countwords.py books/sierra.txt sierra.datUsing Make Wildcards
The Make % wildcard can only be used in a target and in
its dependencies. It cannot be used in actions. In actions, you may
however use $*, which will be replaced by the stem with
which the rule matched.
Our Makefile is now much shorter and cleaner:
# Generate summary table.
results.txt : testzipf.py isles.dat abyss.dat last.dat
python $^ > $@
# Count words.
.PHONY : dats
dats : isles.dat abyss.dat last.dat
%.dat : countwords.py books/%.txt
python $^ $@
.PHONY : clean
clean :
rm -f *.dat
rm -f results.txt- Use the wildcard
%as a placeholder in targets and dependencies. - Use the special variable
$*to refer to matching sets of files in actions.
Content from Variables
Last updated on 2023-04-24 | Edit this page
Estimated time: 20 minutes
Overview
Questions
- How can I eliminate redundancy in my Makefiles?
Objectives
- Use variables in a Makefile.
- Explain the benefits of decoupling configuration from computation.
Despite our efforts, our Makefile still has repeated content, i.e.
the name of our script – countwords.py, and the program we
use to run it – python. If we renamed our script we’d have
to update our Makefile in multiple places.
We can introduce a Make variable (called a macro in some versions of Make) to hold our script name:
COUNT_SRC=countwords.pyThis is a variable assignment
- COUNT_SRC is assigned the value
countwords.py.
We can do the same thing with the interpreter language used to run the script:
LANGUAGE=python$(...) tells Make to replace a variable with its value
when Make is run. This is a variable reference. At any place where we
want to use the value of a variable we have to write it, or reference
it, in this way.
Here we reference the variables LANGUAGE and
COUNT_SRC. This tells Make to replace the variable
LANGUAGE with its value python, and to replace
the variable COUNT_SRC with its value
countwords.py.
Defining the variable LANGUAGE in this way avoids
repeating python in our Makefile, and allows us to easily
change how our script is run (e.g. we might want to use a different
version of Python and need to change python to
python2 – or we might want to rewrite the script using
another language (e.g. switch from Python to R)).
Use Variables
Update Makefile so that the %.dat rule
references the variable COUNT_SRC. Then do the same for the
testzipf.py script and the results.txt rule,
using ZIPF_SRC as the variable name.
This Makefile contains a solution to this challenge.
We place variables at the top of a Makefile so they are easy to find
and modify. Alternatively, we can pull them out into a new file that
just holds variable definitions (i.e. delete them from the original
Makefile). Let us create config.mk:
# Count words script.
LANGUAGE=python
COUNT_SRC=countwords.py
# Test Zipf's rule
ZIPF_SRC=testzipf.pyWe can then import config.mk into Makefile
using:
include config.mkWe can re-run Make to see that everything still works:
We have separated the configuration of our Makefile from its rules –
the parts that do all the work. If we want to change our script name or
how it is executed we just need to edit our configuration file, not our
source code in Makefile. Decoupling code from configuration
in this way is good programming practice, as it promotes more modular,
flexible and reusable code.
- Define variables by assigning values to names.
- Reference variables using
$(...).
Content from Functions
Last updated on 2026-02-17 | Edit this page
Estimated time: 25 minutes
Overview
Questions
- How else can I eliminate redundancy in my Makefiles?
Objectives
- Write Makefiles that use functions to match and transform sets of files.
At this point, we have the following Makefile:
include config.mk
# Generate summary table.
results.txt : $(ZIPF_SRC) isles.dat abyss.dat last.dat
$(LANGUAGE) $^ > $@
# Count words.
.PHONY : dats
dats : isles.dat abyss.dat last.dat
%.dat : $(COUNT_SRC) books/%.txt
$(LANGUAGE) $^ $@
.PHONY : clean
clean :
rm -f *.dat
rm -f results.txtMake has many functions which
can be used to write more complex rules. One example is
wildcard. wildcard gets a list of files
matching some pattern, which we can then save in a variable. So, for
example, we can get a list of all our text files (files ending in
.txt) and save these in a variable by adding this at the
beginning of our makefile:
TXT_FILES=$(wildcard books/*.txt)We can add a .PHONY target and rule to show the
variable’s value:
.PHONY : variables
variables:
@echo TXT_FILES: $(TXT_FILES)@echo
Make prints actions as it executes them. Using @ at the
start of an action tells Make not to print this action. So, by using
@echo instead of echo, we can see the result
of echo (the variable’s value being printed) but not the
echo command itself.
If we run Make:
We get:
OUTPUT
TXT_FILES: books/abyss.txt books/isles.txt books/last.txt books/sierra.txtNote how sierra.txt is now included too.
patsubst (‘pattern substitution’) takes a pattern, a
replacement string and a list of names in that order; each name in the
list that matches the pattern is replaced by the replacement string.
Again, we can save the result in a variable. So, for example, we can
rewrite our list of text files into a list of data files (files ending
in .dat) and save these in a variable:
DAT_FILES=$(patsubst books/%.txt, %.dat, $(TXT_FILES))We can extend variables to show the value of
DAT_FILES too:
.PHONY : variables
variables:
@echo TXT_FILES: $(TXT_FILES)
@echo DAT_FILES: $(DAT_FILES)If we run Make,
then we get:
OUTPUT
TXT_FILES: books/abyss.txt books/isles.txt books/last.txt books/sierra.txt
DAT_FILES: abyss.dat isles.dat last.dat sierra.datNow, sierra.txt is processed too.
With these we can rewrite clean and
dats:
.PHONY : dats
dats : $(DAT_FILES)
.PHONY : clean
clean :
rm -f $(DAT_FILES)
rm -f results.txtLet’s check:
We get:
OUTPUT
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/isles.txt isles.dat
python countwords.py books/last.txt last.dat
python countwords.py books/sierra.txt sierra.datWe can also rewrite results.txt:
results.txt : $(ZIPF_SRC) $(DAT_FILES)
$(LANGUAGE) $^ > $@If we re-run Make:
We get:
OUTPUT
python countwords.py books/abyss.txt abyss.dat
python countwords.py books/isles.txt isles.dat
python countwords.py books/last.txt last.dat
python countwords.py books/sierra.txt sierra.dat
python testzipf.py last.dat isles.dat abyss.dat sierra.dat > results.txtLet’s check the results.txt file:
OUTPUT
Book First Second Ratio
abyss 4044 2807 1.44
isles 3822 2460 1.55
last 12244 5566 2.20
sierra 4242 2469 1.72So the range of the ratios of occurrences of the two most frequent words in our books is indeed around 2, as predicted by Zipf’s Law, i.e., the most frequently-occurring word occurs approximately twice as often as the second most frequent word. Here is our final Makefile:
include config.mk
TXT_FILES=$(wildcard books/*.txt)
DAT_FILES=$(patsubst books/%.txt, %.dat, $(TXT_FILES))
# Generate summary table.
results.txt : $(ZIPF_SRC) $(DAT_FILES)
$(LANGUAGE) $^ > $@
# Count words.
.PHONY : dats
dats : $(DAT_FILES)
%.dat : $(COUNT_SRC) books/%.txt
$(LANGUAGE) $^ $@
.PHONY : clean
clean :
rm -f $(DAT_FILES)
rm -f results.txt
.PHONY : variables
variables:
@echo TXT_FILES: $(TXT_FILES)
@echo DAT_FILES: $(DAT_FILES)Remember, the config.mk file contains:
# Count words script.
LANGUAGE=python
COUNT_SRC=countwords.py
# Test Zipf's rule
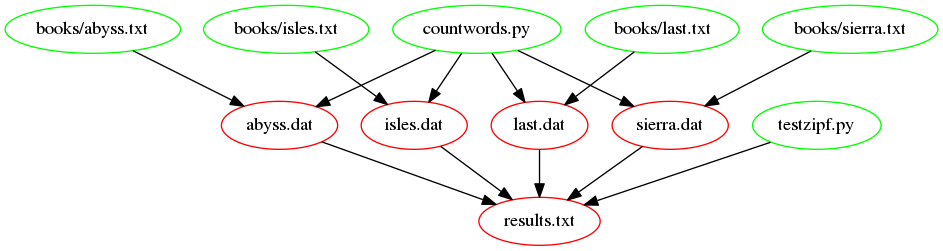
ZIPF_SRC=testzipf.pyThe following figure shows the dependencies embodied within our
Makefile, involved in building the results.txt target, now
we have introduced our function:
Adding more books
We can now do a better job at testing Zipf’s rule by adding more books. The books we have used come from the Project Gutenberg website. Project Gutenberg offers thousands of free ebooks to download.
Exercise instructions:
- go to Project Gutenberg and use the search box to find another book, for example ‘The Picture of Dorian Gray’ from Oscar Wilde.
- download the ‘Plain Text UTF-8’ version and save it to the
booksfolder; choose a short name for the file (that doesn’t include spaces) e.g. “dorian_gray.txt” because the filename is going to be used in theresults.txtfile - optionally, open the file in a text editor and remove extraneous
text at the beginning and end (look for the phrase
END OF THE PROJECT GUTENBERG EBOOK [title]) - run
makeand check that the correct commands are run, given the dependency tree - check the results.txt file to see how this book compares to the others
- Make is actually a small programming language with many built-in functions.
- Use
wildcardfunction to get lists of files matching a pattern. - Use
patsubstfunction to rewrite file names.
Content from Self-Documenting Makefiles
Last updated on 2023-04-24 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- How should I document a Makefile?
Objectives
- Write self-documenting Makefiles with built-in help.
Many bash commands, and programs that people have written that can be
run from within bash, support a --help flag to display more
information on how to use the commands or programs. In this spirit, it
can be useful, both for ourselves and for others, to provide a
help target in our Makefiles. This can provide a summary of
the names of the key targets and what they do, so we don’t need to look
at the Makefile itself unless we want to. For our Makefile, running a
help target might print:
OUTPUT
results.txt : Generate Zipf summary table.
dats : Count words in text files.
clean : Remove auto-generated files.So, how would we implement this? We could write a rule like:
.PHONY : help
help :
@echo "results.txt : Generate Zipf summary table."
@echo "dats : Count words in text files."
@echo "clean : Remove auto-generated files."But every time we add or remove a rule, or change the description of a rule, we would have to update this rule too. It would be better if we could keep the descriptions of the rules by the rules themselves and extract these descriptions automatically.
The bash shell can help us here. It provides a command called sed which stands for
‘stream editor’. sed reads in some text, does some
filtering, and writes out the filtered text.
So, we could write comments for our rules, and mark them up in a way
which sed can detect. Since Make uses # for
comments, we can use ## for comments that describe what a
rule does and that we want sed to detect. For example:
## results.txt : Generate Zipf summary table.
results.txt : $(ZIPF_SRC) $(DAT_FILES)
$(LANGUAGE) $^ > $@
## dats : Count words in text files.
.PHONY : dats
dats : $(DAT_FILES)
%.dat : $(COUNT_SRC) books/%.txt
$(LANGUAGE) $^ $@
## clean : Remove auto-generated files.
.PHONY : clean
clean :
rm -f $(DAT_FILES)
rm -f results.txt
## variables : Print variables.
.PHONY : variables
variables:
@echo TXT_FILES: $(TXT_FILES)
@echo DAT_FILES: $(DAT_FILES)We use ## so we can distinguish between comments that we
want sed to automatically filter, and other comments that
may describe what other rules do, or that describe variables.
We can then write a help target that applies
sed to our Makefile:
.PHONY : help
help : Makefile
@sed -n 's/^##//p' $<This rule depends upon the Makefile itself. It runs sed
on the first dependency of the rule, which is our Makefile, and tells
sed to get all the lines that begin with ##,
which sed then prints for us.
If we now run
we get:
OUTPUT
results.txt : Generate Zipf summary table.
dats : Count words in text files.
clean : Remove auto-generated files.
variables : Print variables.If we add, change or remove a target or rule, we now only need to
remember to add, update or remove a comment next to the rule. So long as
we respect our convention of using ## for such comments,
then our help rule will take care of detecting these
comments and printing them for us.
- Document Makefiles by adding specially-formatted comments and a target to extract and format them.
Content from Conclusion
Last updated on 2023-04-24 | Edit this page
Estimated time: 35 minutes
Overview
Questions
- What are the advantages and disadvantages of using tools like Make?
Objectives
- Understand advantages of automated build tools such as Make.
Automated build tools such as Make can help us in a number of ways. They help us to automate repetitive commands, hence saving us time and reducing the likelihood of errors compared with running these commands manually.
They can also save time by ensuring that automatically-generated artifacts (such as data files or plots) are only recreated when the files that were used to create these have changed in some way.
Through their notion of targets, dependencies, and actions, they serve as a form of documentation, recording dependencies between code, scripts, tools, configurations, raw data, derived data, plots, and papers.
Creating PNGs
Add new rules, update existing rules, and add new variables to:
- Create
.pngfiles from.datfiles usingplotcounts.py. - Remove all auto-generated files (
.dat,.png,results.txt).
Finally, many Makefiles define a default phony target called
all as first target, that will build what the Makefile has
been written to build (e.g. in our case, the .png files and
the results.txt file). As others may assume your Makefile
conforms to convention and supports an all target, add an
all target to your Makefile (Hint: this rule has the
results.txt file and the .png files as
dependencies, but no actions). With that in place, instead of running
make results.txt, you should now run make all,
or just simply make. By default, make runs the
first target it finds in the Makefile, in this case your new
all target.
This
Makefile and this
config.mk contain a solution to this challenge.
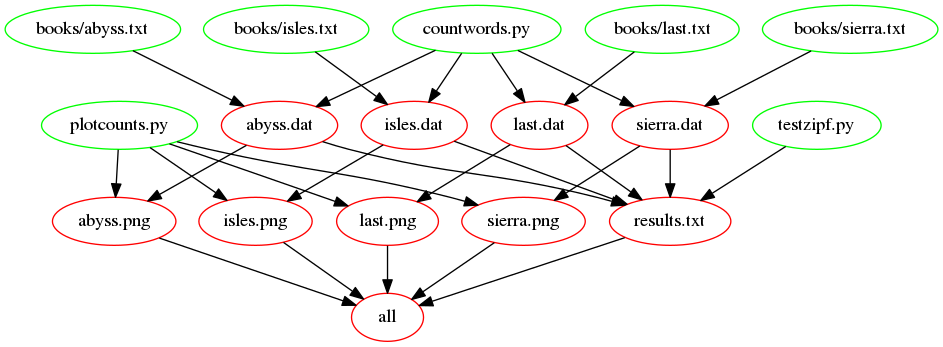
The following figure shows the dependencies involved in building the
all target, once we’ve added support for images:
Creating an Archive
Often it is useful to create an archive file of your project that includes all data, code and results. An archive file can package many files into a single file that can easily be downloaded and shared with collaborators. We can add steps to create the archive file inside the Makefile itself so it’s easy to update our archive file as the project changes.
Edit the Makefile to create an archive file of your project. Add new rules, update existing rules and add new variables to:
Create a new directory called
zipf_analysisin the project directory.-
Copy all our code, data, plots, the Zipf summary table, the Makefile and config.mk to this directory. The
cp -rcommand can be used to copy files and directories into the newzipf_analysisdirectory: Hint: create a new variable for the
booksdirectory so that it can be copied to the newzipf_analysisdirectory-
Create an archive,
zipf_analysis.tar.gz, of this directory. The bash commandtarcan be used, as follows: Update the target
allso that it createszipf_analysis.tar.gz.Remove
zipf_analysis.tar.gzwhenmake cleanis called.Print the values of any additional variables you have defined when
make variablesis called.
This
Makefile and this
config.mk contain a solution to this challenge.
Archiving the Makefile
Why does the Makefile rule for the archive directory add the Makefile to our archive of code, data, plots and Zipf summary table?
Our code files (countwords.py,
plotcounts.py, testzipf.py) implement the
individual parts of our workflow. They allow us to create
.dat files from .txt files, and
results.txt and .png files from
.dat files. Our Makefile, however, documents dependencies
between our code, raw data, derived data, and plots, as well as
implementing our workflow as a whole. config.mk contains
configuration information for our Makefile, so it must be archived
too.
touch the Archive Directory
Why does the Makefile rule for the archive directory
touch the archive directory after moving our code, data,
plots and summary table into it?
A directory’s timestamp is not automatically updated when files are
copied into it. If the code, data, plots, and summary table are updated
and copied into the archive directory, the archive directory’s timestamp
must be updated with touch so that the rule that makes
zipf_analysis.tar.gz knows to run again; without this
touch, zipf_analysis.tar.gz will only be
created the first time the rule is run and will not be updated on
subsequent runs even if the contents of the archive directory have
changed.
- Makefiles save time by automating repetitive work, and save thinking by documenting how to reproduce results.
